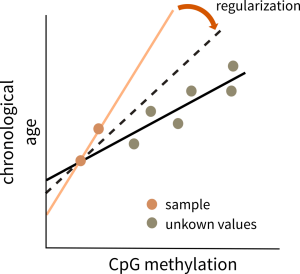
Regularization methods like the elastic-net regression help adjust the weights of methylation signals, facilitating the chronological age prediction.
Graphic: Steve HoffmannPrincipal Investigator: Steve Hoffmann
The Computational Biology Group at the Fritz Lipmann Institute on AgingExternal link (FLI) headed by Steve Hoffmann focuses on the disentanglement of networks governing genome activity during aging and disease. The groups’ expertise in the development of computational methods for epigenomics and transcriptomics is complemented by its wet-lab activities in the field of transcription factor-dependent genome regulation and the role of epigenomic modifications. The groups’ primary interest is the investigation into epigenomic signals and mechanisms relevant for the (dys-)regulation of genome activity in aging and age-related diseases. The group focuses on two closely linked aspects of genome regulation, i.e., consequences of transcription factor activities and their relation to chemical DNA modifications such as cytosine methylation (5mC) and hydroxy-methylation (5hmC). The team investigates this interplay in bottom-up as well as top-down approaches.
Several studies have shown that chronological age can be predicted from the methylation levels of a small number of cytosines. If a test person deviates from the predictions in a particular way, this could indicate accelerated or decelerated biological aging. However, previously established methylation clocks do not differentiate between cytosine methylation (5mC) and the functionally different hydroxymethylation (5hmC). This modification is a promising candidate for a causal link with aging because of age- and disease-related changes in the 5hmC-associated metabolic processes. As part of this work package, we will collect experimental data in parallel for the 5mC and 5hmC modifications, train 5hmC models for age determination, compare them with conventional clocks and carry out a functional classification. A central problem in researching age-related changes is the very definition of biological age. We will therefore compare the 5mC and 5hmC clocks with objective fitness markers. The “fitness watches” developed here primarily enable functional elucidation of age-related epigenomic changes. In addition, the working group contributes its bioinformatics expertise in the integration of high-dimensional molecular and clinical data sets and works closely with the partners of the Adult-LIFE study.
Postdoc: Maja Olecka